HRDP v7.1 in Phenogen Browser
8/23/24 - v3.9.6 - Genome Transcriptome Browser has HRDP v7.1 data
HRDP v7.1 Data Available:
- HRDP v7.1 Tracks: Transcriptome, Splice Junction Tracks
- Coming soon: expression, eQTLs, and WGCNA - HRDP v6(rn7) is still available simply select the HRDP version to display.
HRDP v7 in Phenogen Browser
5/14/24 - v3.9.5 - Genome Transcriptome Browser has HRDP v7 data
HRDP v7 Data Available:
- HRDP v7 Tracks: Transcriptome, Splice Junction Tracks
- Expression: TPM summaries, detailed normalized expression at the region or gene level
- Coming soon: eQTLs and WGCNA - HRDP v6(rn7) is still available simply select the HRDP version to display.
HRDP v7 IsoSeq Data
4/11/24 - IsoSeq data available raw and rn7.2 aligned
IsoSeq data updated:
- Brain/Liver/Merged IsoSeq Tracks showing IsoSeq Read alignments to rn7.
- Created IsoSeq UCSC Genome Browser track hub
- Fixed a gene display bug in the genome browser
- Downloads Added (RNA-Seq Downloads):
- IsoSeq samples high quality Isoform FASTQ files.
- Rn7.2 merged genome alignment of FASTQ files
- Rn7.2 merged transcriptome as GTF
HRDP v7 UCSC Track Hubs
4/3/24 - HRDP v7 UCSC Genome Browser track hubs are available now. Downloads -> UCSC.
- Tissue specific transcriptomes and merged transcriptomes
- Tissue/strain specific read count tracks both total(sum of 3 replicates) or sampled( randomly sampled reads to match the strain with the fewest reads)
- Merged across strain read counts and then sampled to represent 5 Billion reads.
- Splice junctions from merged and 5 billion read sampled Bam files.
- Tissue/strain specific read count tracks both total(sum of 3 replicates) or sampled( randomly sampled reads to match the strain with the fewest reads)
- Merged across strain read counts and then sampled to represent 5 Billion reads.
- Splice junctions from merged and 5 billion read sampled Bam files.
TPM/Heritability Summaries on Gene Reports
2/14/24 - First of many new display functions and refinements. TPM and Heritability across tissues are summarized for each transcript in gene reports. We've included a number of bug fixes. Additional new features are coming.
HRDP v6/v7 TPM Data
PhenoGen HRDP v6/v7 TPM Data is available via the REST API and Website in the downloads section. We continue to work on HRDP v7 data including 73 strains of the HRDP.
UCSC Track Hubs HRDPv6
PhenoGen HRDP v6 UCSC Track Hubs are available now to access Transcriptomes and read depth tracks on UCSC Genome Browser. Downloads -> UCSC
CTC Phenogen Poster
PhenoGen HRDP v6 and REST API updates are highlighted on our CTC poster. Download it here.
Complex Trait Community/Rat Genomics Annual Meeting
9/29/22-9/30/22 Denver,CO / University of Colorado Anschutz Medical Campus
Learn More
Register
Come and see the PhenoGen Poster on the Rn7/HRDP v6 Dataset and the REST API. Download PDF
Learn More
Register
Come and see the PhenoGen Poster on the Rn7/HRDP v6 Dataset and the REST API. Download PDF
Rn7 is available
9/22/22 - Rn7/HRDPv6 is available in the genome browser. Downloads/REST API update soon. WGCNA and eQTLs are not yet available but will be updated soon.
HRDP v6 includes 58 strains in Brain/Liver. Kidney on HXB/BXH is available. Heart with 1 sample in 20 strains of HXB/BXH is also now available.
HRDP v6 includes 58 strains in Brain/Liver. Kidney on HXB/BXH is available. Heart with 1 sample in 20 strains of HXB/BXH is also now available.
REST API
6/22/22 - The dataset functions on the REST API are available to get files for the current datasets in rn6. Rn7 is coming soon.
R functions are also available to pull data directly into R.
R functions are also available to pull data directly into R.
OSGA Webinar #26
Genome-wide Association Study Summary Statistics - Where to find them and how to use them
4/22/22 10am PDT/1pm EDTFree Registeration Here
Summary of this webinar:
This presentation will guide attendees with how to access genome-wide association study summary statistics and showcase resources available for annotating these summary data for follow-up analyses, including gene-based analyses, eQTL, and epigenetic annotation as well as causal variable analysis. We will guide attendees through components of a GWAS summary dataset and two excellent resources - FUMA and MASSIVE - that use these summary files as inputs to generate vast amounts of annotations that can be brought forward to answer translational research questions.
Presented by:
Dr. Arpana Agrawal and Dr. Alexander S. Hatoum
Department of Psychiatry
Washington University School of Medicine
Series Information and Previously Recorded Webinars
OSGA Webinar #25
Mouse Phenome Database: Resources and analysis tools for curated and integrated primary mouse phenotype and genotype data
4/8/22 10am PDT/1pm EDTView Here
Summary of this webinar:
The Mouse Phenome Database (MPD; https://phenome.jax.org) is a widely used resource that provides access to primary experimental data, protocols, and analysis tools for mouse phenotyping studies. Data are contributed by investigators around the world and represent a broad scope of phenotyping endpoints and disease-related characteristics in naïve mice and those exposed to drugs, environmental agents, or other treatments. MPD is engineered to facilitate interactive data exploration and quantitative analysis. It encompasses data from inbred strains and other reproducible panels, including HMDP, KOMP, Collaborative Cross (CC), CC-RIX, and founder strains, along with primary data from mapping populations, including historic mapping crosses and advanced high-diversity mouse populations such as Diversity Outbred mice. A new Study Intake Platform (SIP) for data contributors allows domain experts to submit and annotate their own data with relevant ontology terms. Data contributors also provide detailed information for protocols and animal environmental conditions to fulfill ARRIVE guidelines. Data are exposed to analysis tools within MPD and are available through APIs to other systems. We will demonstrate selected MPD tools, including GenomeMUSter (a new imputed SNP grid on 500+ strains of mice, 83+M locations) and a GWAS metanalysis tool.
Presented by:
Molly Bogue and Robyn Ball
With other senior members of the MPD team: Elissa Chesler, Vivek Philip, Dave Walton
The Jackson Laboratory
Series Information and Previously Recorded Webinars
eQTLs for HRDP v5
11/1/21 - Brain/Liver/Kidney eQTLs using the full panel of HRDP animals available 43 strains have now been calculated with GEMMA and are available now.
OSGA Webinar #23
Julia: a fast, friendly, and powerful language for data science
11/12/21 10am PDT/1pm EDTView Here
Summary of this webinar:
Julia is a high-level dynamic programming language that is gaining popularity. The Julia language is designed for scientific computing and offers several attractive features for data science applications. In this webinar, we will make a case for why a data scientist might consider taking a serious look at Julia. We will show code examples and point the audience to further resources.
- To articulate why Julia is attractive for data scientists
- To provide an overview of Julia language syntax and design
- To provide additional resources about the Julia language and ecosystem
Gregory Farage, PhD
Post-Doctoral Fellow
Department of Preventive Medicine
University of Tennessee Health Science Center
Saunak Sen, PhD
Professor and Division Chief
Department of Preventive Medicine
University of Tennessee Health Science Center
Series Information and Previously Recorded Webinars
Kidney RNA-Seq
9/7/21 - Kidney transcriptome and HXB/BXH expression values and eQTLs are avaialble in the genome browser. WGCNA results will be available soon.
OSGA Webinar #22
Guide to evaluating the application of machine learning methods in genetics literature
10/22/21 10am PDT/1pm EDTView Here
Summary of this webinar:
- To describe the relationship between artificial intelligence (AI), machine learning (ML), and deep learning (DL).
- To describe general scenarios when ML is appropriate.
- To understand methods for comparing the performance of different ML algorithms
- To layout general criteria to examine when evaluating literature that includes machine learning algorithms
Laura Saba, PhD
Associate Professor
Department of Pharmaceutical Sciences
University of Colorado Anschutz Medical Campus
Series Information and Previously Recorded Webinars
PhenoGen update 3.7.5
12/11/20 update to v3.7.5
- Sampled counts from every strain are now available as well as total counts.
- You can create a custom view and select total/sampled count and choos the density of the count tracks.
- Bug Fixes
PhenoGen update 3.7.4
11/18/20 update to v3.7.4
- You can now create gene lists from Genome Browser tables - Filter tables from tracks, eQTLs, WGCNA based on expression/tissue and then use gene list tools on the list.
- Implemented a reasonable region restriction of 5Mbp for now. - We are working to create a less detailed view that will be displayed in larger regions.
- Bug Fixes
PhenoGen v3.7.3
- Version 3.7.3 has been released with the following updates.
- RNA-Seq eQTLs based on HRDP
- Heritability based on HRDP
- TPM summary
- Track Table filtering
- Table export to clipboard, csv, xls, and PDF
- Ensembl and Reconstruction based eQTLs/TPM/Heritability
PhenoGen v3.7.2
- Version 3.7.2 has been released with the following updates.
- RNA-Seq eQTLs
- Gene Track Summaries now include RNA-Seq based eQTL summaries
- Modules for a region can be selected base on overlap of the module QTL with the region.
- Genes with an eQTL that overlaps the region are now available based on RNA-Seq and Microarrays
- Custom Views
- Total Counts for all 45 strains to date are available.
- Read counts for all 45 strains are available to add as browser tracks.
- Bug Fixes
- RNA-Seq eQTLs
PhenoGen v3.7.1
- Version 3.7.1 has been released with the following updates.
- Downloads
- Tissue Specific/Merged Reconstruction GTFs
- Normalized Expression Values for Ensembl/Reconstruction, Gene/Transcript, Strain Means/Individual values
- Custom Views
- Build a custom view on the fly by selecting a few check boxes simply select sissue and track types.
- Views can be named and saved for future use
- Downloads
PhenoGen v3.7
- Version 3.7 has been released with HRDP v5.
- Transcriptomes are now based on the full HXB/BXH RI panel in brain and liver and available inbred strains now including the F344/Stm and LE/Stm strains.
- Updated WGCNA, expression values, eQTLs, and transcriptomes are available.
- Transcript level expression is available for the full panel.
- Transcript level eQTLs are now available once you've selected a gene.
- Transcript level WGCNA is coming in the next 1-2 months.
- CircularRNA data is availble now. This includes predictions from BNLx/SHR in Brain/Heart/Liver using CIRI 2.0 and CircExplorer2. Also Heart left ventricle includes data from more than 12,000 previously described cirRNAs using cirRNA arrays. Full reports are coming soon but the browser includes tracks with annotation and structure all available data.
PhenoGen v3.6
- Version 3.6 has been released with HRDP v4. Transcriptomes are now based on the full HXB/BXH RI panel in brain and liver and available inbred strains except for the F344/Stm and LE/Stm. The FXLE/LEXF parents will be included in HRDP v5. Updated WGCNA, expression values, eQTLs, and transcriptomes are available. Transcript level WGCNA is coming in the next 1-2 months. Transcript level expression is available again now for the full panel.
- Links to Literature searches for gene-gene/gene-keyword relationshipts and gene-Addiction relationships.
- Shortcuts to the genome browser to get you to sections of interest more directly.
- New gene list eQTL image/browser.
Rat Genomics Workshop 8/2-8/3
Look for another workshop spring/summer 2020
Summer of Rat Genomics and Genetics Workshop - Aug 2nd-3rd, 2019 - Colorado - NIDA Center for Omics, Systems Genetics and the AddictomeCome to Colorado in August for a 2-day hands-on workshop on web-based tools for rat genetics/genomics including RGD, GeneNetwork2, PhenoGen, GeneWeaver, and several others. Offered by the NIDA Center for Omics, Systems Genetics, and the Addictiome.
More Info
Registration
Complex Traits Consortium 2019
PDFs of posters for CTC 2019 Meeting:
Circular RNA Array/Predictions
PhenoGen REST API - gene level data
Circular RNA Array/Predictions
PhenoGen REST API - gene level data
Phenogen in the Cloud
This is the new home for PhenoGen running in the cloud. As such, a large number of changes have occurred in the background that will help us
maintain
the site and improve performance. Please excuse some ongoing bugs as we are beta testing the site now. This is not currently the recommended
version.
However, we would appreciate any help testing the supported features by simply using this site as you would the previous site.
REST API Survey 2/20/2018
Please help us prioritize effort on a REST API to provide access to data on Phenogen. Take the 1 question survey below.
PhenoGen v3.4.2 3/9/2018
Added read depth count tracks to the genome browser for whole brain and liver for the inbred strains:
ACI, Dark-Agouti, Cop, F344-NCl, F344-NHsd, LEW-Crl, LEW-SsNHsd, SHRSP, SR-JrHsd, SS-JrHsd, and WKY.
PhenoGen v3.4.1 2/16/2018
The Genome/Transcriptome Data Browser can now look up genes by either their gene or transcript PhenoGen ID.
NIDA Genetics Consortium Meeting Poster
NIDA Meeting Poster -
Download the poster from the NIDA meeting with an outline of ways to use our WGCNA Modules and recent examples of our use with phenotype QTLs.
Download here.
PhenoGen v3.4 12/10/2017
Recombinant Inbred Small RNA - Added expression data for small RNA features across RI Panel in Whole
Brain
and
Liver.
PhenoGen v3.3 4/30/2017
Recombinant Inbred Total RNA - Added expression data for reconstructed transcripts across RI Panel in
Whole
Brain and Liver. Added RNA-Seq based WGCNA for Whole Brain and Liver.
PhenoGen v3.2 11/13/2016
Small RNA - added tracks and detail on all known and novel (predicted by MiRDeep and SNOSeeker) small
RNAs
in
Brain, Heart, and Liver from the BNLx/SHR parental strains.
Merged Total RNA Transcriptome - added a track with the merged transcriptome from the 3 available tissues and assigned new unique PhenoGen IDs to all novel transcripts.
Merged Total RNA Transcriptome - added a track with the merged transcriptome from the 3 available tissues and assigned new unique PhenoGen IDs to all novel transcripts.
PhenoGen v3.1 6/15/2016
Rn6 - is available in the browser, for gene list analysis, and both RNA-Seq datasets and microarray
datasets
have been updated.
Rat WGCNA - Heart and Liver have been added to the rn6 data.
Rat WGCNA - Heart and Liver have been added to the rn6 data.
PhenoGen v3.0 5/31/2016
Anonymous Gene List - You can now use our gene list analysis tools without registering. We encourage
you to
link your email so you don't loose access to previous work.
Rn6 (June 2016): Rn6 - Will be available in June 2016. Rn6 and Rn5 will be available in the genome browser, public HXB datasets, and gene list analysis tools.
Rn6 (June 2016): Rn6 - Will be available in June 2016. Rn6 and Rn5 will be available in the genome browser, public HXB datasets, and gene list analysis tools.
PhenoGen v2.16.1 11/10/2015
Security Updates - We now require using HTTPS so all of the data transmitted between your browser and
our
server is encrypted. Update your bookmarks.
Future Update: Rn6 - We are still working on updating the Microarrays and RNA-Seq data to Rn6. Our next major update will include Rn6.
Future Update: Rn6 - We are still working on updating the Microarrays and RNA-Seq data to Rn6. Our next major update will include Rn6.
PhenoGen v2.16 7/21/2015
- Gene List tabs have been reformatted so you can submit submit new analyses and view results and running analyses status from a single page.
- GO term summaries are available for Gene Lists.
- MuliMiR results are available for Rat both in Gene Lists and the browser(individual genes, WGCNA modules).
Minor Updates 6/8/2015
We've made a couple of minor updates since the last release.
- The UCSC Repeat Mask track is now available.
- You now have the ability to add tracks without going into the edit view option.
- We've also fixed a number of bugs and made general functionality improvements.
Workshop Video/Slides 4/16/2015
v2.15 of PhenoGen 3/7/2015


We've added GO term summary and miRNA targeting views to the Weighted Gene Co-expression Network Analysis. Look at what's new for a summary of changes.
HTTPS support 2/9/2015
We now support https to keep your connections more secure. We will eventually redirect all traffic to the secure site,
but for now feel free to try it out here: https://phenogen.ucdenver.edu/PhenoGen/
v2.14 of PhenoGen 1/10/2015
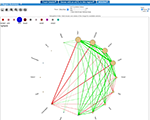
We've added Weighted Gene Co-expression Network Analysis. Look at what's new for a summary of changes.
v2.13 of PhenoGen 9/27/2014
We've updated PhenoGen. Look at what's new for a summary of changes.
Added multiMiR
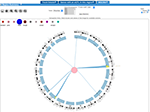
Using multiMiR(an R package available here) you can view validated and predicted miRNAs that target specific genes. You can also select a miRNA and view all genes targeted by the miRNA. multiMiR is avaialble as a new tab for a selected gene in the Genome/Transcriptome Data Browser and in Gene Lists after selecting a list. It is currently available only for mouse genes, but will be available in rat soon.
Added Rat Liver Transcriptome
We've added rat liver tracks including, a transcriptome reconstructiong track, splice junction track, and stranded read depth count tracks.
Available in
the Genome/Transcriptome Browser.
Follow on Facebook/Google+/Twitter
Follow PhenoGen to keep up with new features, demonstrations, and help by providing feedback to direct future updates.
RNA-Seq Data Summary Graphics
Rat Brain RNA-Seq data summary graphics are now available. Click below to browse the RNA-Seq data summary:
Reconstructed transcripts from this RNA-Seq data are still combined with PhenoGen array data in Genome/Transcriptome Data Browser.
Reconstructed transcripts from this RNA-Seq data are still combined with PhenoGen array data in Genome/Transcriptome Data Browser.

